What is PCR ?
 Introduction:
Introduction:
PCR is Polymerase Chain Reaction (PCR) is a molecular biology technique used to amplify specific DNA sequences. Making millions of copies of a particular segment. It is essential in various applications, including genetic research, diagnostics, and forensic analysis.
Types of PCR
- Standard PCR
The original method, used to amplify DNA fragments through repeated cycles of denaturation, annealing, and extension.
- Real-Time PCR (qPCR)
Allows for the quantification of DNA as it amplifies, using fluorescent dyes to monitor the process in real-time.
- Reverse Transcription PCR (RT-PCR)
Single strand RNA into double stranded DNA using reverse transcriptase before amplification, commonly used for gene expression studies.
- Nested PCR
Involves two sets of primers and two rounds of amplification, increasing specificity and sensitivity for detecting low-abundance targets.
- Multiplex PCR
Amplifies multiple DNA targets in a single reaction using multiple primer sets, enabling simultaneous detection of different sequences.
- Digital PCR (dPCR)
Provides absolute quantification of DNA by partitioning the sample into many reactions, allowing for precise measurements.
- Hot Start PCR
Uses modified DNA polymerases that are inactive at lower temperatures to prevent non-specific amplification, improving specificity.
- Touchdown PCR
Gradually decreases the annealing temperature in the initial cycles to enhance specificity before continuing with standard conditions.
Ingredients Used in PCR
- Template DNA
– The DNA that has the specific sequence we want to make more copies of.
- Primers
Short, single-stranded sequences of nucleotides that are complementary to the target DNA regions, essential for initiating the amplification process.
- DNA Polymerase
An enzyme that creates new DNA strands by adding nucleotides that match the existing template strand. Common examples of these enzymes are Taq polymerase and Pfu polymerase.
- Nucleotides (dNTPs)
The building blocks of DNA, including deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), deoxy guanosine triphosphate (dGTP), and deoxythymidine triphosphate (dTTP).
- Buffer Solution
Provides the optimal pH and ionic environment for the PCR reaction, typically containing Tris-HCl and KCl.
- MgCl₂ (Magnesium Chloride)
A cofactor that is essential for the activity of DNA polymerase and helps stabilize the DNA structure during amplification.
Method involves in PCR
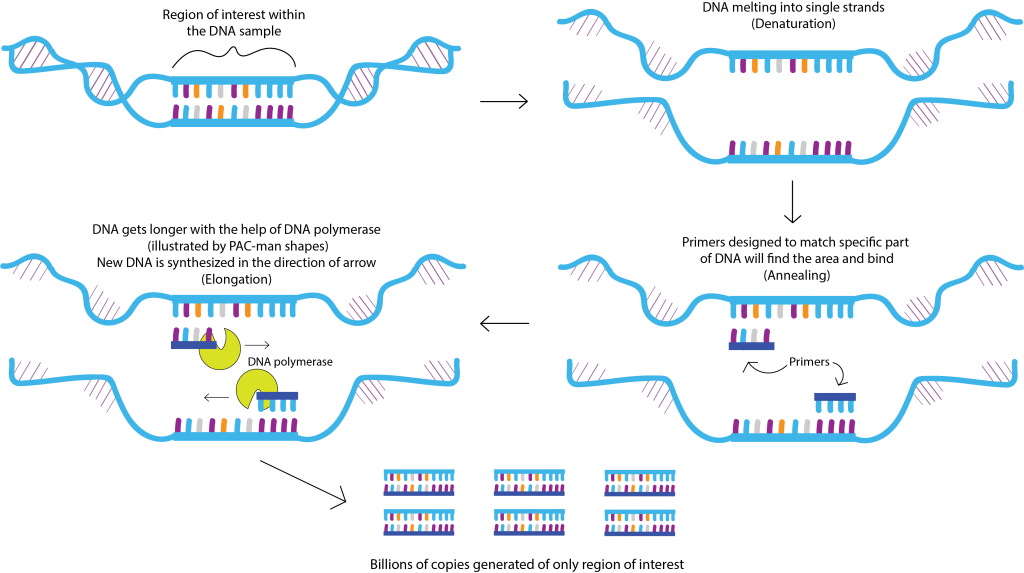
Polymerase Chain Reaction (PCR) involves a series of well-defined steps that facilitate the amplification of a specific DNA segment. The typical PCR process consists of three main stages: denaturation, annealing, and extension. Here’s a detailed breakdown of the method:
- Preparation of Reaction Mix
Combine the following components in a PCR tube:
Template DNA: The DNA containing the target sequence.
Primers: Forward and reverse primers specific to the target region.
DNA Polymerase: A thermostable enzyme (e.g., Taq polymerase).
dNTPs: A mix of deoxyadenosine, deoxycytidine, deoxyguanosine, and deoxythymidine triphosphates.
Buffer: Provides the necessary pH and ions (often Tris-HCl and KCl).
MgCl₂: Essential for DNA polymerase activity.
- Denaturation
Temperature: Typically, around 94-98°C.
The reaction mixture is heated to separate the double-stranded DNA into single strands. This denaturation step usually lasts for 20-30 seconds.
- Annealing
Temperature: Typically, 50-65°C, depending on primer sequences.
The temperature is reduced so that the short pieces of DNA called primers can attach themselves to the matching parts of the single-stranded DNA. This process typically takes about 20 to 30 seconds.
- Extension
Temperature: Generally, around 72°C.
The DNA polymerase synthesizes a new DNA strand by adding dNTPs to the 3′ end of the annealed primers. This step typically lasts for 30 seconds to several minutes, depending on the length of the target DNA.
- Cycling
The process involves three main steps (denaturation, annealing, extension) that are repeated for 25 to 40 times. Each time this process is repeated, the amount of the specific DNA we want doubles, resulting in a rapid increase in its quantity.
6.Final Extension
After cycling, a final extension step at 72°C for 5-10 minutes may be performed to ensure that all DNA strands are fully extended.
- Cooling
The reaction is cooled to 4°C for short-term storage of the PCR products.
Visualization and Analysis
After amplification, the PCR products can be analyzed using gel electrophoresis to confirm the presence and size of the amplified DNA.
Errors in PCR
While Polymerase Chain Reaction (PCR) is a powerful and widely used technique, it is not without its potential pitfalls. Errors can occur at various stages of the PCR process, leading to suboptimal results. Here are some common errors and their causes:
1.Non-Specific Amplification
Cause: Occurs when primers bind to sequences other than the intended target, leading to the amplification of undesired products.
Solutions: Optimize primer design to ensure specificity.
Adjust annealing temperature to enhance binding fidelity.
- Primer-Dimer Formation
Cause: Primers can bind to each other instead of the target DNA, forming unwanted amplification products.
Solutions: Design primers that do not have complementary regions. Use a higher annealing temperature to reduce primer-dimer formation.
- Insufficient Template DNA
Cause: Low amounts of target DNA can lead to weak or no amplification.
Solutions: Increase the amount of template DNA used in the reaction.
4.Inhibitors in the Reaction
Cause: Contaminants (e.g., proteins, phenols, or alcohols) can inhibit the activity of DNA polymerase.
Solutions: Purify the DNA sample to remove inhibitors. Use a more robust DNA polymerase that can tolerate inhibitors.
5.Inaccurate Temperature Cycling
Cause: Incorrect cycling temperatures can lead to inefficient amplification or complete failure.
Solutions: Ensure accurate calibration of the thermocycler. Double-check the cycling parameters.
- Suboptimal MgCl₂ Concentration
Cause: Too little or too much magnesium can affect DNA polymerase activity and amplification efficiency.
Solutions: Optimize MgCl₂ concentration through systematic testing.
7.Poor Primer Design
Cause: Primers that are too short, too long, or have a high GC content can lead to inefficient amplification.
Solutions: Follow best practices for primer design, ensuring appropriate length and melting temperature (Tm).
- Contamination
Cause: Introduction of foreign DNA can lead to non-specific amplification or false positives.
Solutions: Use aseptic techniques and dedicated equipment. Include negative controls to detect contamination.
- Over cycling
Cause: Too many amplification cycles can lead to the amplification of non-specific products or degrade specific products.
Solutions: Determine the optimal number of cycles through preliminary experiments.
- Storage and Handling Errors
Cause: Improper storage of reagents (e.g., primers, enzymes) can lead to degradation.
Solutions: Store reagents according to manufacturer guidelines and avoid repeated freeze-thaw cycles.
